Efficient Sequence Alignment Technique for Bioinformatics
Bioinformatics research field is becoming an increasingly important field of research. The reason for that is the increasing interest in studying the structure and the function of DNA, and correlating this information with diseases.
Overview
Bioinformatics research field is becoming an increasingly important field of research. The reason for that is the increasing interest in studying the structure and the function of DNA, and correlating this information with diseases.
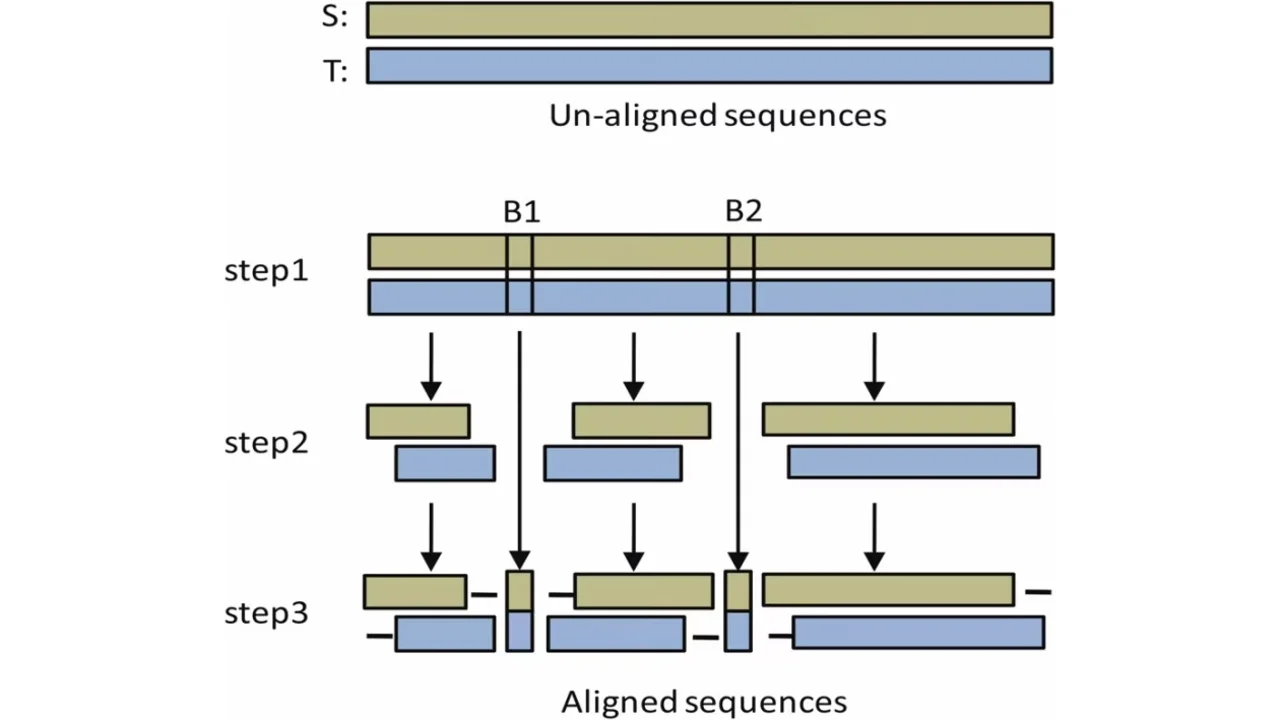
DNA and Protein database sequence alignment such as Smith-Waterman Algorithm is one of the most important applications in Bioinformatics. This algorithm guarantees to return the optimal alignment of two sequences. The first one is called Query and the second one is called Database. These applications need to process large amounts of data which may reach more than 11 GB size and that may take hours of mainframe time to get the optimum solution.
PIs: Dr. Talal Bonny and M Affan Zidan
References:
- T. Bonny and K.N. Salama, Database Filtering Technique for High Speed Database Applications,IEEE Engineering in Medicine and Biology Conference (EMBC'12), submitted
- T. Bonny and K.N. Salama, “Fast Global Sequence Alignment Algorithm,” IEEE Asilomar conference on signals, systems and computers, pp. 1046-1049, 2011
- M. Affan Zidan, Talal Bonny, and K. N. Salama, “High Performance Technique for Database Applications Using Hybrid GPU/CPU Platform,”IEEE Great Lakes Symposium on VLSI (GLSVLSI), pp. 85-90, Lausanne, Switzerland, May 2011.
- Talal Bonny, M. Affan. Zidan, and K. N. Salama, “An Adaptive Hybrid Multiprocessor Technique for Bioinformatics Sequence Alignment,” The 5th International Biomedical Engineering Conference in Cairo (CIBEC), pp.112-115, December 2010, Cairo, Egypt.